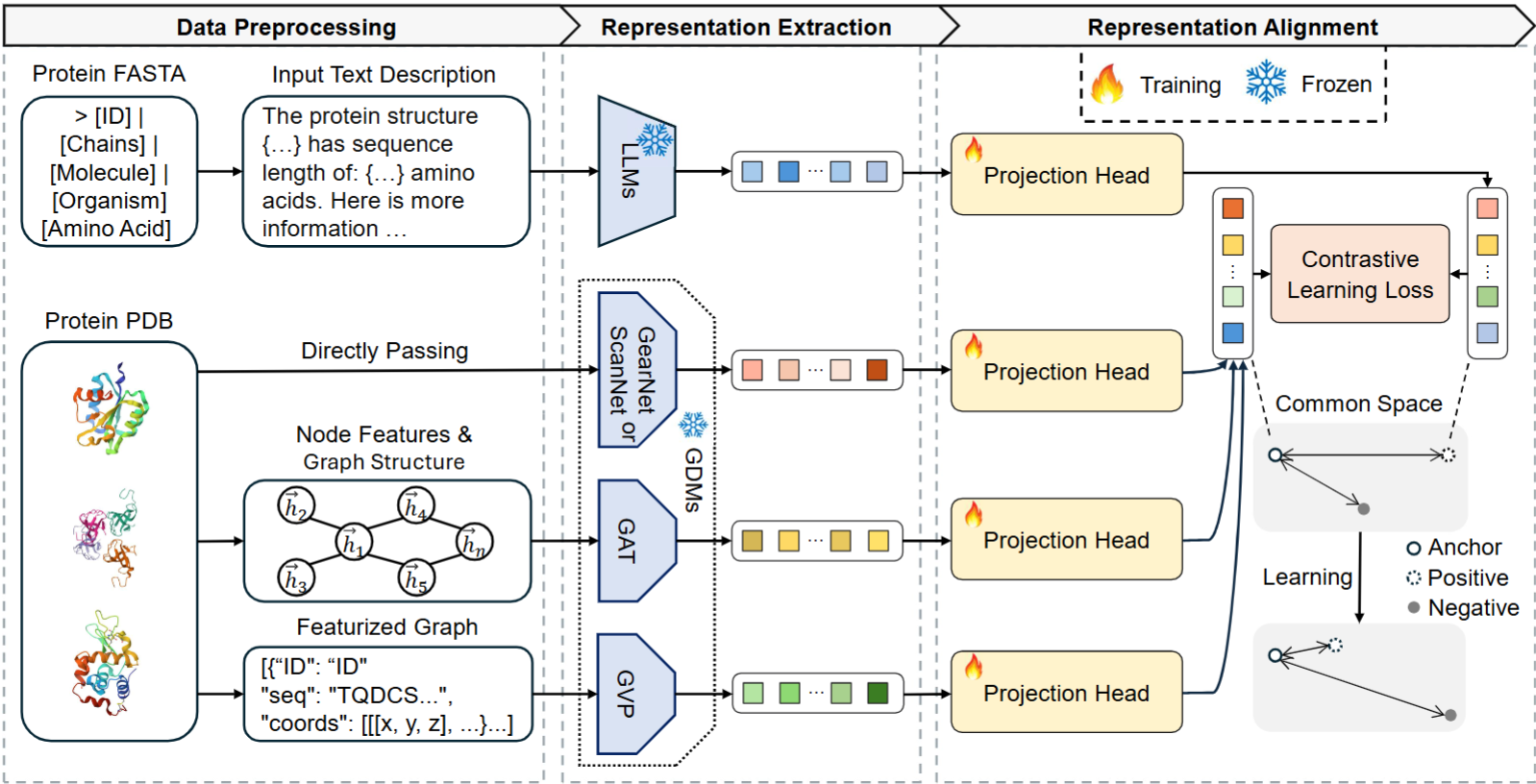

Latent representation alignment has become a foundational technique for constructing multimodal large language models (MLLM) by mapping embeddings from different modalities into a shared space, often aligned with the embedding space of large language models (LLMs) to enable effective cross-modal understanding. While preliminary protein-focused MLLMs have emerged, they have predominantly relied on heuristic approaches, lacking a fundamental understanding of optimal alignment practices across representations. In this study, we explore the alignment of multimodal representations between LLMs and Geometric Deep Models (GDMs) in the protein domain. We comprehensively evaluate three state-of-the-art LLMs (Gemma2-2B, LLaMa3.1-8B, and LLaMa3.1-70B) with four protein-specialized GDMs (GearNet, GVP, ScanNet, GAT). Our work examines alignment factors from both model and protein perspectives, identifying challenges in current alignment methodologies and proposing strategies to improve the alignment process. Our key findings reveal that GDMs incorporating both graph and 3D structural information align better with LLMs, larger LLMs demonstrate improved alignment capabilities, and protein rarity significantly impacts alignment performance. We also find that increasing GDM embedding dimensions, using two-layer projection heads, and fine-tuning LLMs on protein-specific data substantially enhance alignment quality. These strategies offer potential enhancements to the performance of protein-related multimodal models.
Keywords: MultiModal AI, AI for Science, Large Language Models, Explainable AI
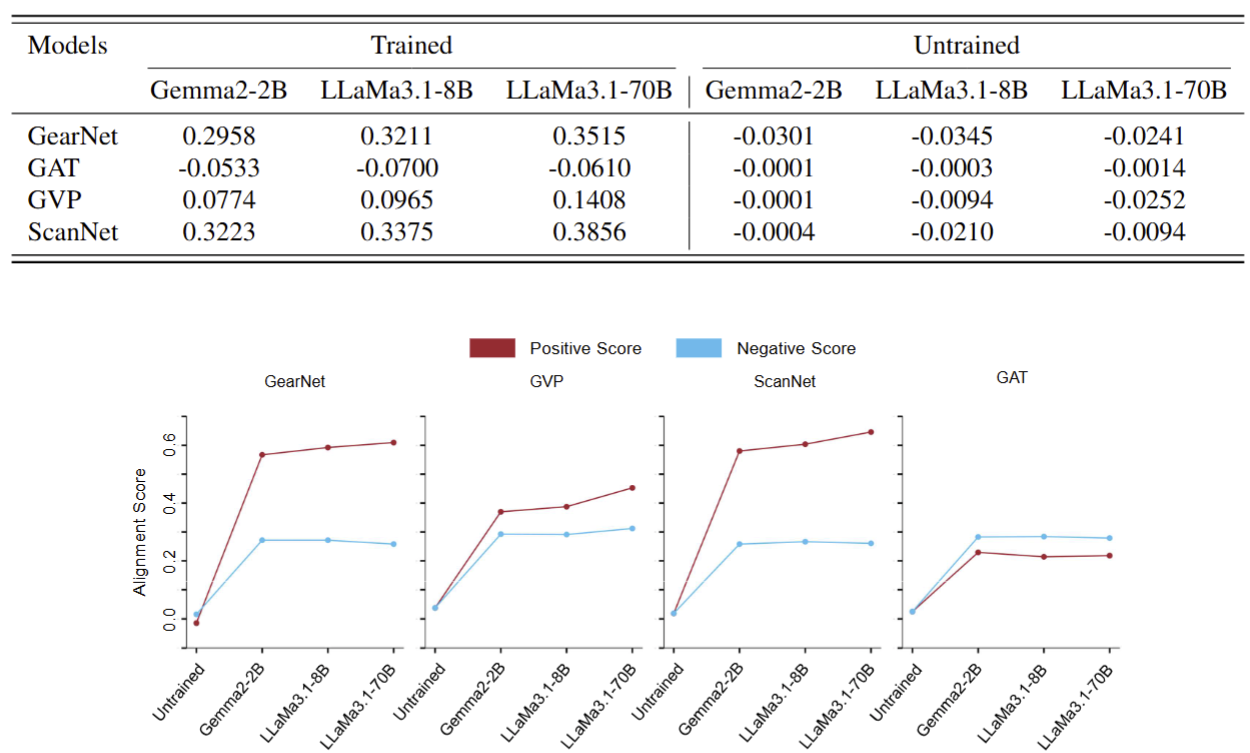
Which LLM-GDM model pairs demonstrate the best alignment performance? We explored which pairs of LLMs and GDMs achieve better alignment. We found that GDMs capable of learning not only from the graph structure of proteins but also from their 3D geometric information tend to align more effectively with LLMs. Additionally, LLMs with higher embedding dimensions consistently demonstrated better alignment with GDMs.

Is there a correlation between different model pairs? We observed that model pairs with strong alignment tend to have higher correlation with other high-performing model pairs.
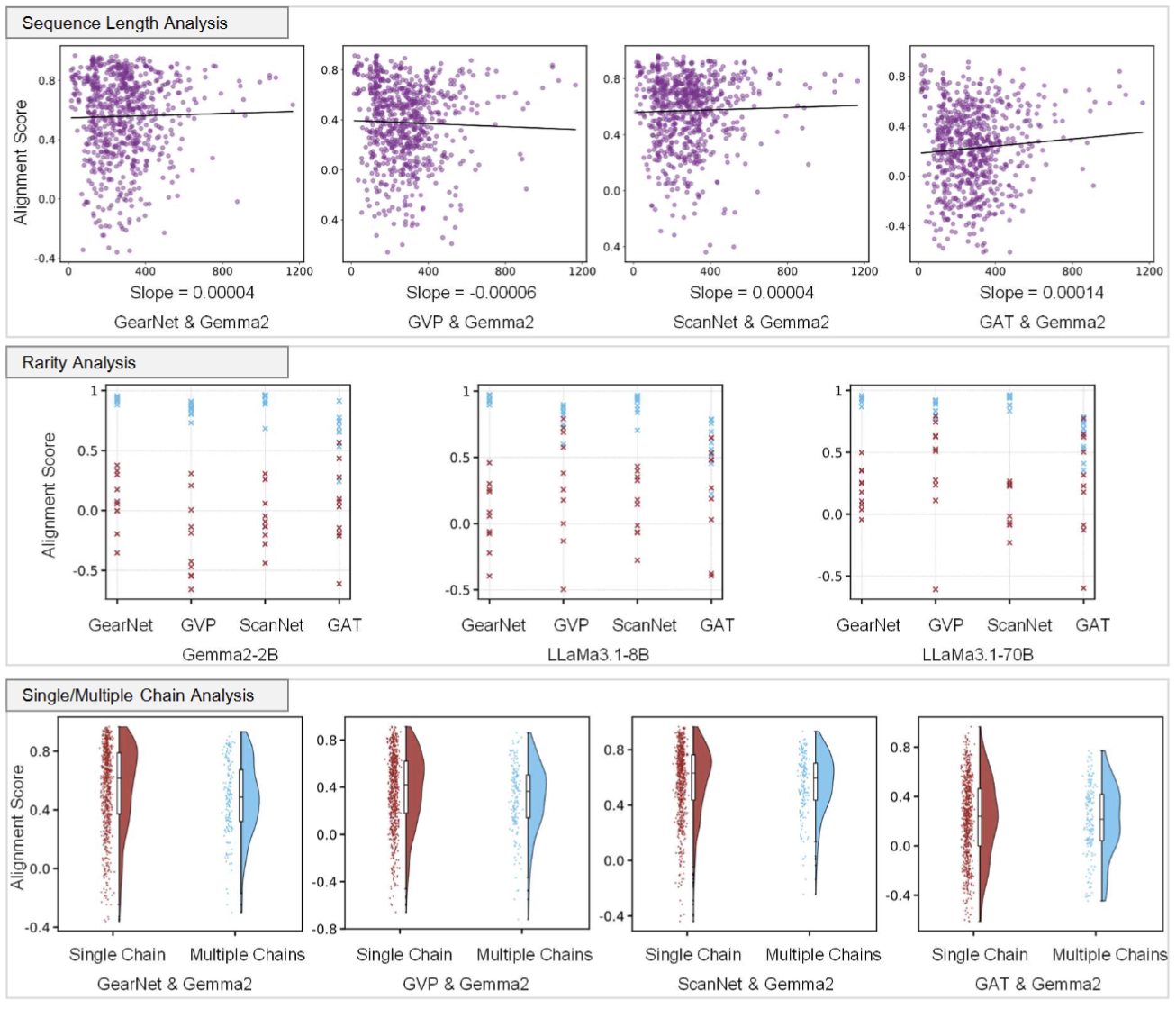
What types of proteins align well across all model pairs, and which do not? We discovered that popular proteins are generally easier to align across model pairs, whereas rare proteins present greater challenges in achieving alignment.
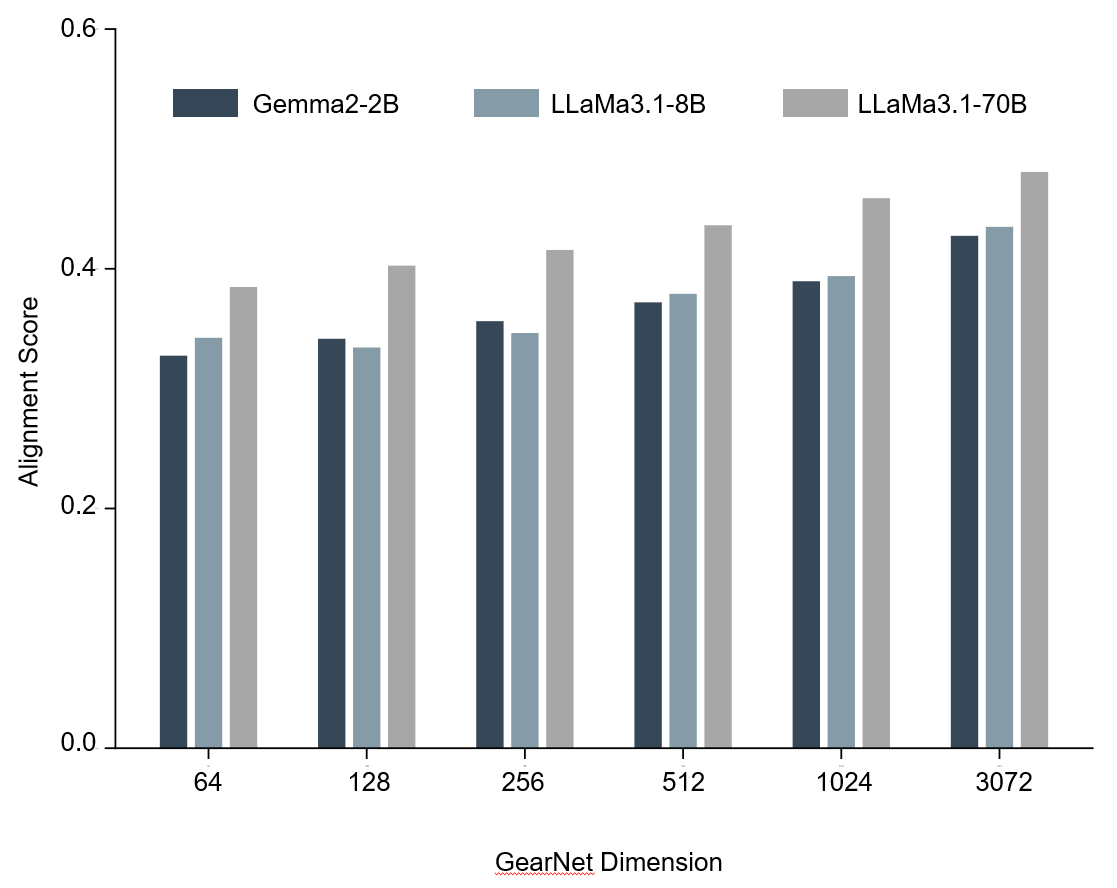
Does increasing the GDM dimension improve alignment performance? Our findings indicate that increasing the embedding dimension size of a GDM enhances its alignment with LLMs.
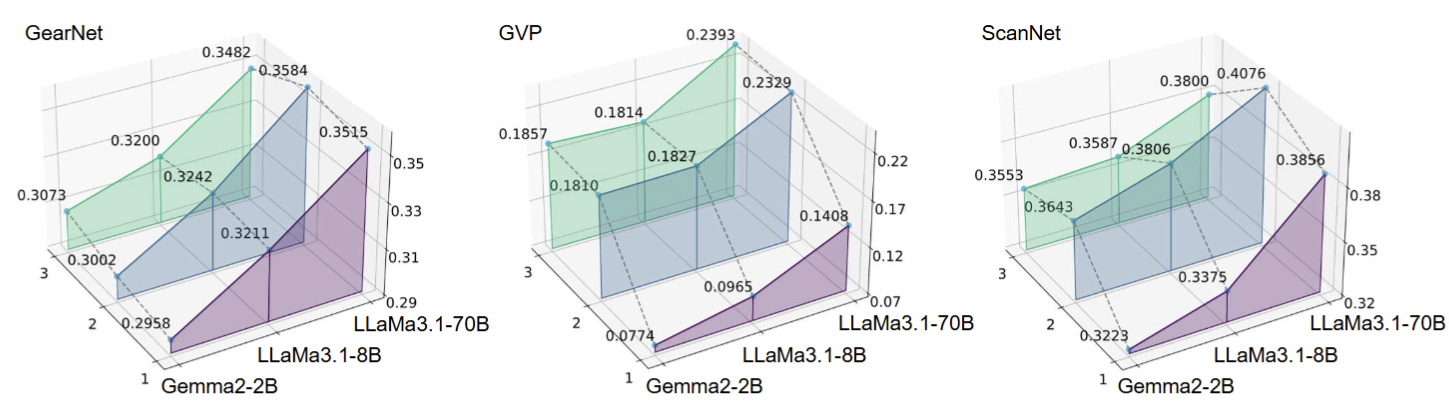
Does adding layers to the GDM's projection head enhance alignment performance? Adding an extra linear layer to the GDM's projection head can significantly improve alignment performance, though this improvement stopped after two layers, suggesting diminishing returns with further complexity.
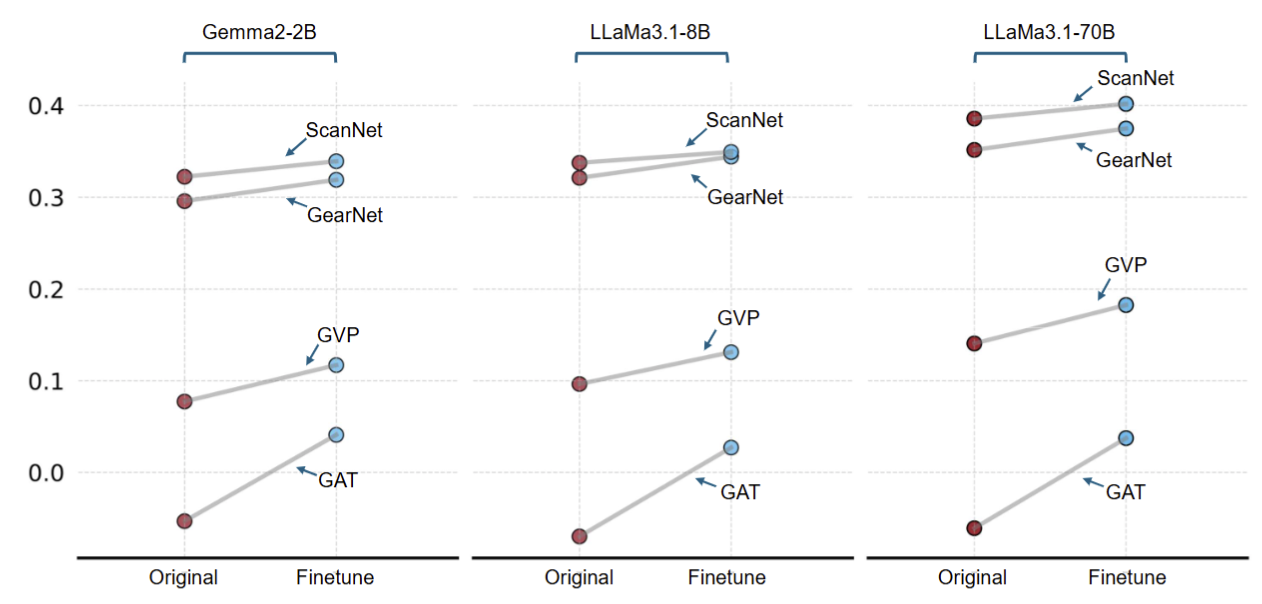
Does fine-tuning an LLM on protein data enhance alignment performance? Another key insight is that fine-tuning LLMs with protein-specific knowledge can lead to better alignment with GDMs, highlighting the value of domain-specific adaptation.
@misc{shu2024exploringalignmentlandscapellms,
title={Exploring the Alignment Landscape: LLMs and Geometric Deep Models in Protein Representation},
author={Dong Shu and Bingbing Duan and Kai Guo and Kaixiong Zhou and Jiliang Tang and Mengnan Du},
year={2024},
eprint={2411.05316},
archivePrefix={arXiv},
primaryClass={cs.LG},
url={https://arxiv.org/abs/2411.05316},
}